AI reveals how a new antibiotic affects gut bacteria.
AI maps how a new antibiotic targets gut bacteria | MIT News
Understanding the Challenges of Antibiotics for Inflammatory Bowel Disease
For patients suffering from inflammatory bowel disease (IBD), traditional antibiotics can often feel like a double-edged sword. While these broad-spectrum medications are frequently prescribed to manage gut flare-ups, they can inadvertently harm beneficial microbes in the intestine. This collateral damage sometimes exacerbates symptoms over time, leading to complications. In this context, bringing a heavy-handed approach to a nuanced problem like gut inflammation may not be ideal.
The Emergence of Enterololin
Researchers at the Massachusetts Institute of Technology’s (MIT) Computer Science and Artificial Intelligence Laboratory (CSAIL) and McMaster University have developed a groundbreaking compound named enterololin. This novel molecule adopts a more targeted strategy for combatting bacteria associated with Crohn’s disease flare-ups, sparing the rest of the microbiome from disruption.
Utilizing a generative AI model, the research team accelerated the mapping of how enterololin functions—a task that typically requires years of work was completed in just months. “This discovery addresses a significant challenge in antibiotic development,” states Jon Stokes, the senior author of the study and an assistant professor at McMaster University.
The Role of AI in Drug Development
One of the major hurdles in antibiotic development has been understanding the exact mechanisms through which these compounds operate within bacterial systems. It’s not enough to identify molecules that kill bacteria in a laboratory; a comprehensive understanding of their effects within living organisms is essential for creating safe, effective therapies.
Enterololin: A Step Toward Precision Antibiotics
Enterololin represents a significant advancement in the field of precision antibiotics. In preclinical models mimicking Crohn’s-like inflammation, this drug specifically targeted Escherichia coli, a problematic gut bacterium known to aggravate flare-ups, while leaving most other microbial agents unharmed. Mice treated with enterololin showed quicker recovery times and maintained healthier microbial communities compared to those given vancomycin, a commonly prescribed antibiotic.
Discovering the Mechanism of Action
Determining a drug’s mechanism of action—the specific molecular target it interacts with—generally requires extensive experimental work. However, Stokes’ lab utilized a high-throughput screening method for the initial discovery of enterololin. The next challenge was addressing the bottleneck of identifying its target.
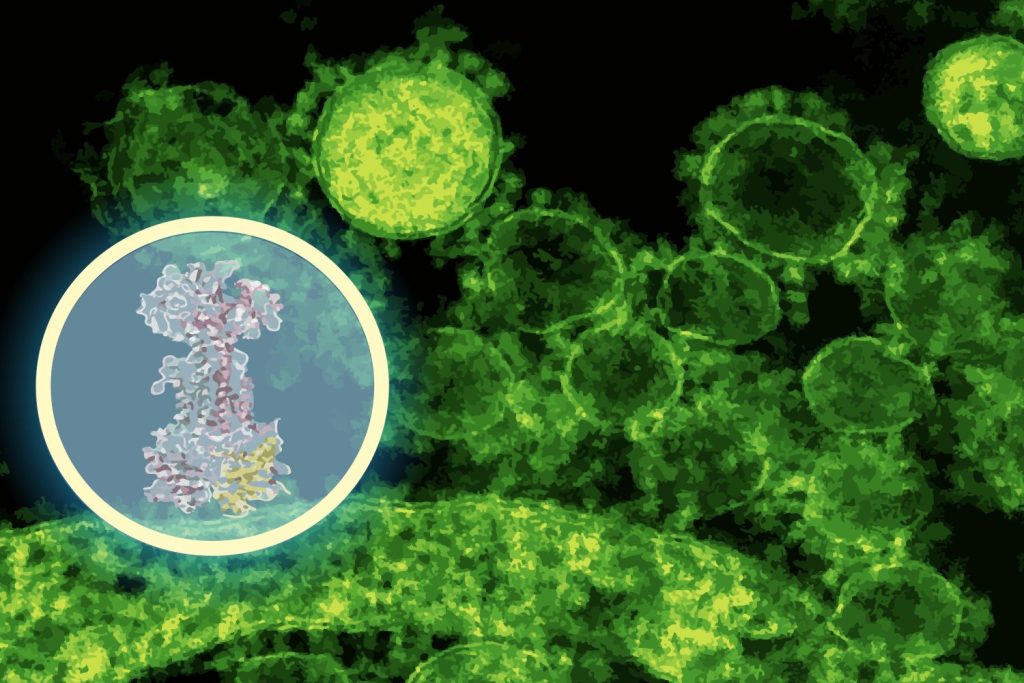
For this, the team leveraged DiffDock, a generative AI model developed by CSAIL in collaboration with MIT researcher Gabriele Corso and Professor Regina Barzilay. This AI system overcomes the limitations of traditional docking algorithms, reframing the problem in terms of probabilistic reasoning. Instead of merely searching through possible configurations, DiffDock iteratively refines guesses to identify the most probable binding site.
Validation Through Experimentation
In a matter of minutes, DiffDock suggested that enterololin binds to a protein complex called LolCDE, which plays a crucial role in transporting lipoproteins in certain bacteria. According to Barzilay, this prediction provided a substantial lead to guide further experiments.
To validate this model, Stokes’ group created enterololin-resistant mutants of E. coli, which indicated genetic changes aligned with the predicted binding site. They also performed RNA sequencing to observe gene expression alterations upon drug exposure and employed CRISPR technology to selectively knock down the expression of the anticipated target. These experiments confirmed disruptions in pathways associated with lipoprotein transport, aligning with DiffDock’s predictions.
“When computational models and laboratory data converge on the same mechanism, that’s when you start to feel you’ve made a breakthrough,” notes Stokes.
The Impact of AI on Drug Discovery
The findings from this study shed light on a transformative approach for utilizing AI in life sciences. While existing AI applications in drug discovery often focus on searching through chemical spaces for active molecules, this study illustrates that AI can also provide essential mechanistic insights. These insights are critical for moving a new compound through the development pipeline.
Mechanism-of-action studies frequently represent significant bottlenecks in drug development, traditionally taking 18 months to two years and costing millions. Surprisingly, the MIT-McMaster team managed to streamline this process, reducing the timeline to around six months and doing so at a fraction of the cost.
Future Prospects for Enterololin
Although still in the preliminary stages of development, enterololin is on a promising path. Stokes’ spinout company, Stoked Bio, has licensed the compound and is currently optimizing its properties for potential human use. Moreover, early research is underway to explore derivatives of enterololin against other resistant pathogens, such as Klebsiella pneumoniae. If successful, clinical trials could commence within the next few years.
Broader Implications for Antibiotic Development
The significance of narrow-spectrum antibiotics has long been highlighted, particularly in terms of minimizing collateral damage to the microbiome. However, discovering and validating such formulations has proven to be a Herculean task. AI tools like DiffDock can make this quest considerably more feasible, opening avenues for a new generation of targeted antimicrobials.
For patients with Crohn’s or other inflammatory bowel conditions, the potential for a drug that alleviates symptoms without adversely affecting the microbiome could dramatically enhance the quality of life. In the broader context, the advent of precision antibiotics could contribute significantly to addressing the pressing issue of antimicrobial resistance.
Conclusion
“What excites me is the prospect of understanding the mechanisms of action in a more efficient manner by blending AI, human intuition, and laboratory research,” Stokes concludes. This approach could revolutionize drug discovery not just for Crohn’s disease but for a variety of illnesses.
As Yves Brun, a professor at the University of Montreal and an expert in antimicrobial resistance, mentions, “AI is becoming an essential tool in our battle against resistant bacteria.” This study exemplifies the innovative applications of AI to elucidate the mechanisms of new antibiotic candidates—a crucial step on the journey toward viable therapeutic developments.
Collaborative Efforts and Future Research
The research included contributions from various McMaster University scientists and was supported by multiple funding agencies, emphasizing the collaborative nature of scientific inquiry. The sequencing data from this research has been made publicly available, along with the release of the DiffDock-L code on GitHub, promoting further innovation in the field.
As we continue to grapple with the complexities of antibiotic resistance, advancements like enterololin coupled with AI’s growing influence could provide hope for more effective, targeted treatments in the future.
Thanks for reading. Please let us know your thoughts and ideas in the comment section down below.
Source link
#maps #antibiotic #targets #gut #bacteria #MIT #News